AI-Designed mutation Resistant Antibodies against Multiple SARS-CoV-2 Strains
Yue Kang (1), Yang Jiao (1) , Cong Chen (2) , Paige Vinson (3), Lynn Rasmussen (3), Lurong Pan* (1)
Introduction
- The emerging variants including Delta and Omicron may not retain activity with current available mAb products that were developed during the initial outbreak with the B.1 strain.
- In this study, we have applied AI technologies in the computational design of broad neutralization antibodies against over 1300 different historical SARS-CoV-2 strains with very small computational cost.
- These AI-designed antibodies also exhibited a very high cross-binding hit rate against different Spike protein RBD region mutants in ELISA assay.
- The AI-designed antibodies were tested in vitro and demonstrated high potency against multiple strains in real virus neutralization assays
Methods and Data
- In the study of SARS-Cov-2, 1300 different historical SARSCoV-2 strains data was collected from based on the GISAID[1][2][3] database as of August 26, 2021
- The first round of AI-based antibody design was performed on selected antibody templates[4][5] in order to increase binding affinities with respect to over 1300 different historical SARSCoV-2 strains including wildtype(B.1) strain and Delta strain. Our large molecule virtual screening platform was able to search 1010 somatic mutation space and 50 cross-binding antibody sequences were designed across different strains.
- The second round antibody design was performed during the prevalence of Omicron strain, in which same computational approach was employed with Omicron strain included. 20 cross-binding antibody sequences were designed.
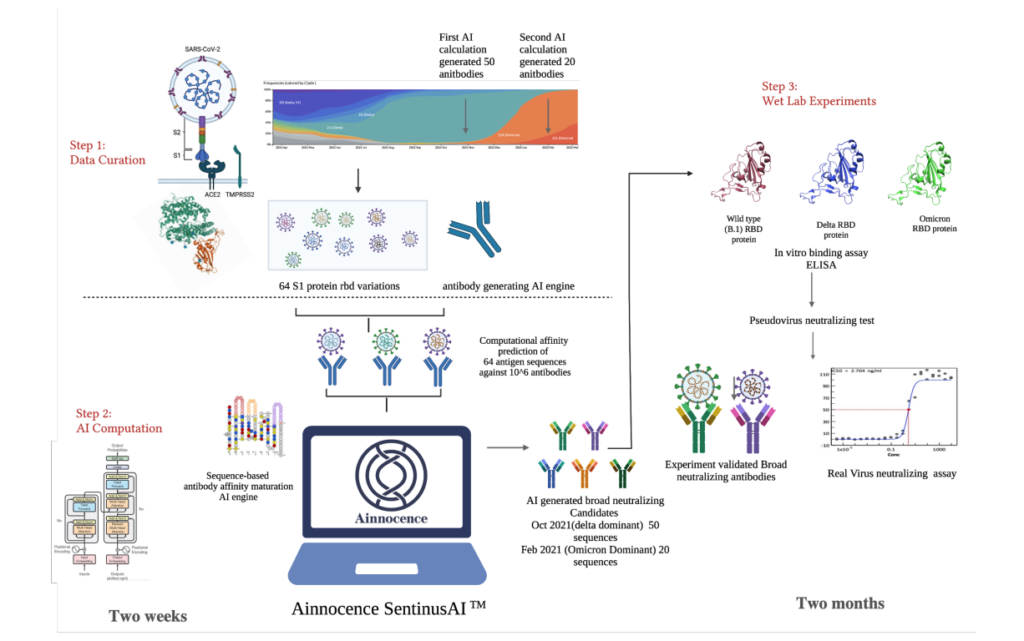
Experiment Results
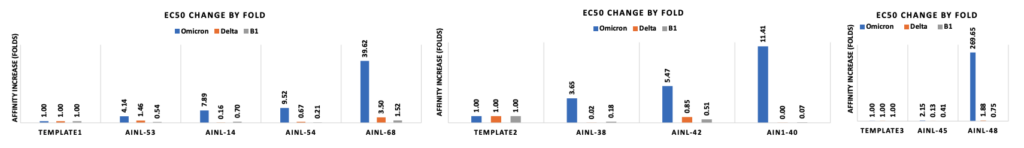
The first batch of 50 antibodies and second batch of 20 antibodies are well-expressed with high hit rate on positive affinity against B.1, Delta and Omicron. The two batches yield 14% and 40% triple cross binding hit rates respectively. Affinity maturation against Omicron on three different template antibodies were optimized up to 269.65 fold increase.
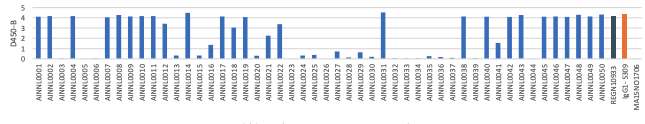
(A) Wildtype RBD Protein
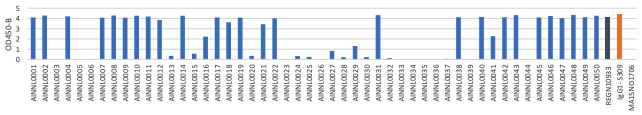
(B) Delta RBD Protein
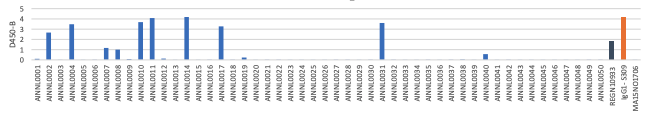
(C) Omnicron RBD Protein
A. OD 450 absorbance values obtained by direct ELISA of 50 antibodies (1μg/ml; blue) against wild type (A), delta (B) and Omicron(C) at 1ug/ml. Plate coated with 1ug/ml RBD protein. OD450 = optical density at 450 nm; Two therapeutic SARS-CoV-2 antibodies were tested as controls (black,orange)
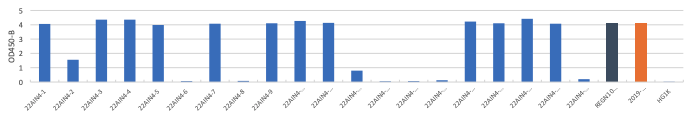
(A) Wildtype RBD Protein
(B) Delta RBD Protein
(C) Omnicron RBD Protein
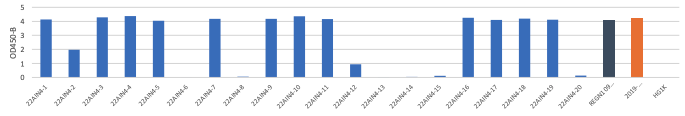
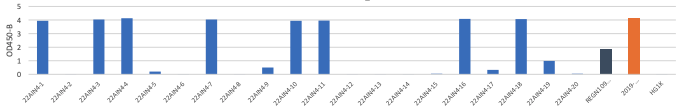
B. OD 450 absorbance values obtained by direct ELISA of 20 antibodies (1μg/ml; blue) against wild type (A), delta (B) and Omicron(C ). Plate coated with 1ug/ml RBD protein. OD450 = optical density at 450 nm; Two therapeutic SARS-CoV-2 antibodies were tested as controls (black,orange)
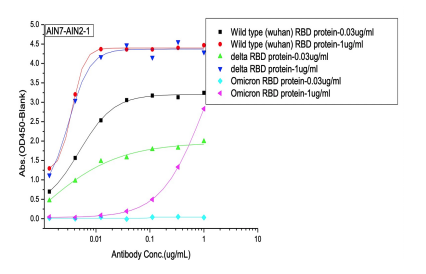
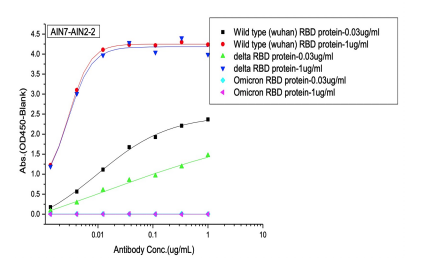
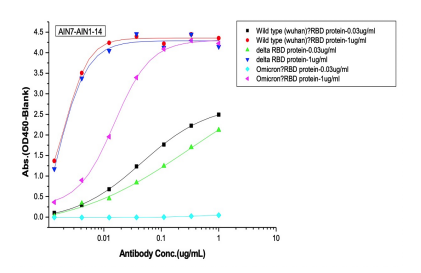
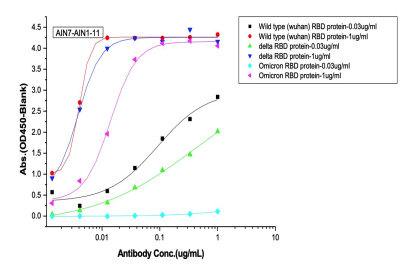
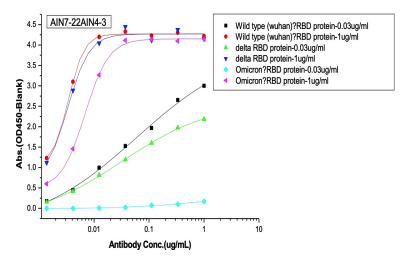
Dose dependent ELISA binding test was also performed on antibodies and controls against Sars-CoV-2 Wild type (B.1), Delta and Omicron RBD proteins to determine the IC50 value.
(A) Controls: approved therapeutic antibodies
(B) 1St batch (pre-omicron) AI-designed cross-binding antibodies
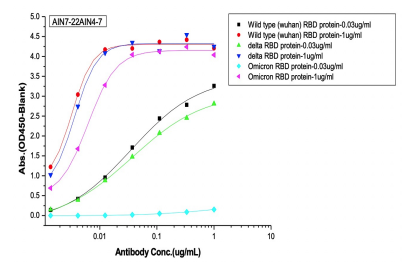
(C) 2nd batch (post-omicron) AI-designed cross-binding antibodies
Selected Elisa dose dependent curves of two different concentrations(0.03ug/ml, 1ug/ml) of RBD (WT, Delta, Omicron) proteins were coated on the plate and 7 gradient concentrations of mAbs added to the plate. (A) Controls of previously approved therapeutic antibodies (B) 1st batch AI designed antibodies before Omicron prevalence (C) 2nd batch AI designed antibodies after Omicron prevalence.
Real Virus Neutralization: the antivirus effect of designed antibodies was evaluated by cell-based assay measuring the cytopathic effect (CPE) of the virus infecting Vero E6 host cells against Delta and Omicron strains. 10 antibodies show neutralization effect on Delta Strain(with IC50 <10ug/ml). 1 antibody shows effect on Omicron Strain
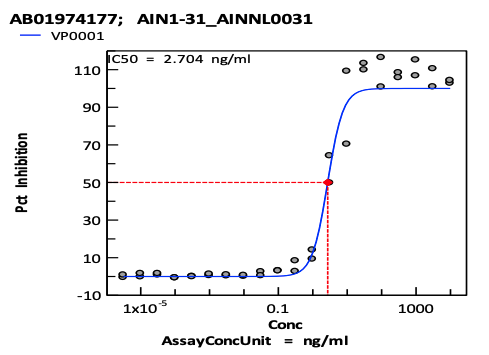
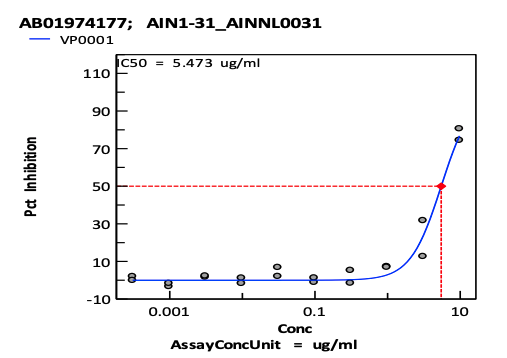
CPE inhibition curves of selected antibodies against Delta(A) and Omicron(B). Neutralization of the virus is measured by incubation of serial dilutions of antibody with infectious virus particles for 2 hrs at 37ºC. Vero E6 cells enriched for ACE2 expression were added and plates incubated for 72 hrs at 37°C/5%CO2 and 90% humidity. Cell Titer-Glo (Promega) was added to each well and luminescence read using a BMG CLARIOstar plate reader to measure cell viability.
Conclusions
The present study demonstrates AI’s capability in antibodies design with in vitro efficacy as well as drastically reduced time and cost of antibody engineering such as affinity maturation. The study indicates that AI can design cross-binding antibodies against many different antigen populations such as viral mutant strains. The proposed modeling hypothesize that AI can learn hidden patterns of a viral evolution process and design antibodies against future virus beyond current strains.
Acknowledgements
This project is supported by Ainnocence. We thank the protein expression and ELISA assay provided by Sino Biological and real virus inhibition assay provided by Southern Research. Ainnocence and Sino Biological are currently under strategic collaboration to provide AI powered antibody discovery service.
